Updating metadata
Using Swagger
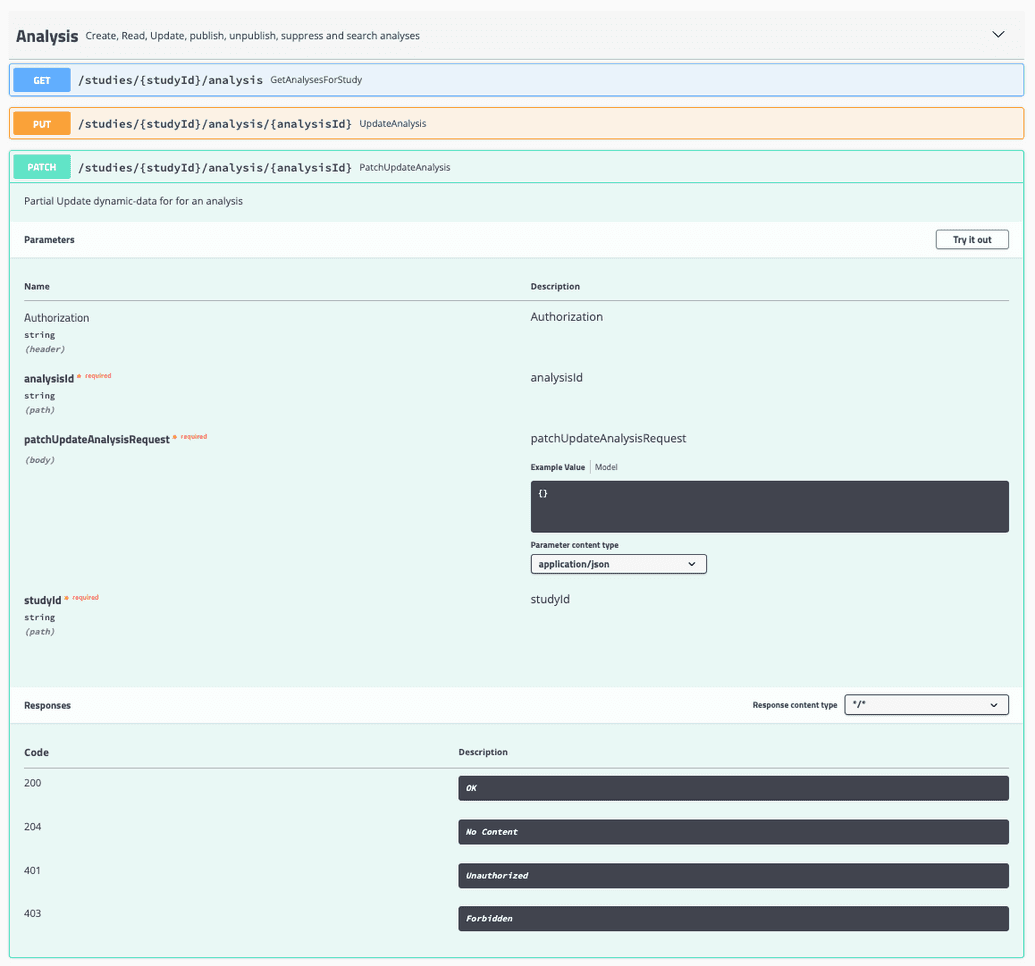
An individual analysis can be updated through the Patch PatchUpdateAnalysis endpoint located in the Analysis dropdown on the Swagger UI.
The PatchUpdateAnalysis endpoint requires the following inputs:
- Your Authorization token
- An analysisID
- The updated Analysis content
- A StudyId
Programmatic Updates
Here, we provide some examples of updating analyses using the Song API programmatically. If you have any questions or require technical support for your specific use case, please contact us on our Slack channel.
Updating an analysis
The following script shows how to update a specific analysis in a given study by making a PATCH request to Song’s API:
### Define variablesanalysis_id="ANALYSIS-ABC123-SAMPLEA"url="https://song.virusseq-dataportal.ca"study="ABC123"api_token="PASSWORD"### PATCH endpointendpoint="%s/studies/%s/analysis/%s" % (url,study,analysis_id)headers={"accept":"*/*","Authorization":"Bearer %s" % (api_token),"Content-Type": "application/json"}payload = {'experiment':{'sequencing_instrument':new_value}}patch_response=requests.patch(endpoint,json=payload,headers=headers)if patch_response.status_code!=200:print("error calling patch endpoint",endpoint,patch_response.status_code)break
Updating multiple analyses
The following script demonstrates how to update multiple analyses associated with specific samples within a study. This is achieved by iterating over a list of sample IDs, retrieving the corresponding analyses, and modifying a specific attribute of each analysis. The script utilizes the PATCH request method to update the analyses in the data portal's API.
### Define variablessamples_to_mod=["SAMPLE_A","SAMPLE_B","SAMPLE_C","SAMPLE_D","SAMPLE_E","SAMPLE_F"]url="https://song.virusseq-dataportal.ca"study="ABC123"api_token="PASSWORD"for sample in samples_to_mod:### Retrieve analyses associated to samplesendpoint="%s/studies/%s/analysis/search/id?submitterDonorId=%s" % (url,study,sample)headers={"accept":"*/*"}get_response=requests.get(endpoint,headers=headers)### Catch endpoint call failif get_response.status_code!=200:print("Error :%s" % (endpoint))break### We only expect 1 analysisif len([analysis for analysis in get_response.json() if analysis['analysisState']=='PUBLISHED'])==0:print("No published detected :%s" % (endpoint))breakif len([analysis for analysis in get_response.json() if analysis['analysisState']=='PUBLISHED'])>1:print("Multiple analsyes detected :%s" % (endpoint))breakanalysis=get_response.json()[0]analysis_id=analysis['analysisId']old_value=get_response.json()[0]['experiment']['sequencing_instrument']new_value="Illumina MiSeq"### PATCH endpointendpoint="%s/studies/%s/analysis/%s" % (url,study,analysis_id)headers={"accept":"*/*","Authorization":"Bearer %s" % (api_token),"Content-Type": "application/json"}payload = {'experiment':{'sequencing_instrument':new_value}}patch_response=requests.patch(endpoint, json=payload,headers=headers)if patch_response.status_code!=200:print("error calling patch endpoint",endpoint,patch_response.status_code)break
 Song-assigned IDs
Song-assigned IDs
Song-assigned IDs (donor, sample, specimen, analysis and object IDs) and those specified in the base schema are immutable and cannot be altered. In the event you need to change any of these values, it is suggested to UNPUBLISH and SUPPRESS the analysis and resubmit the analysis.